
 ## TODOs
- [x] Paper released
- [x] Code released
- [x] Slide-SAM-B weights released
- [x] Slide-SAM-H weights released
## Models
### Large scale Medical Image Pretrained Weights
| name | resolution | Prompt | Weights |
| :---------: | :---------: | :---------: | :----------------------------------------------------------: |
| Slide-SAM-B | 1024 x 1024 | box & point | [ckpt](https://pan.baidu.com/s/1jvJ2W4MK24JdpZLwPqMIfA) [code: 7be9] |
| Slide-SAM-H | 1024 x 1024 | box & point | [ckpt](https://pan.baidu.com/s/1jnOwyWd-M1fBIauNi3IA4w) [code: 05dy] |
## Getting Started
### Install tutils tools
```
pip install trans-utils
```
### Prepare datasets
We recommend you to convert the dataset into the nnUNet format.
```
00_custom_dataset
imagesTr
xxx_0000.nii.gz
...
labelsTr
xxx.nii.gz
...
```
Try to use the function organize in [nnunet-style](https://github.com/MIC-DKFZ/nnUNet/blob/master/documentation/dataset_format.md) or ```organize_by_names``` to prepare your custom datasets.
Then run :
```python
python -m datasets.generate_txt
```
A ```[example]_train.txt``` will be generated in ```./datasets/dataset_list/```
The content should be like below
```
01_BCV-Abdomen/Training/img/img0001.nii.gz 01_BCV-Abdomen/Training/label/label0001.nii.gz
01_BCV-Abdomen/Training/img/img0002.nii.gz 01_BCV-Abdomen/Training/label/label0002.nii.gz
01_BCV-Abdomen/Training/img/img0003.nii.gz 01_BCV-Abdomen/Training/label/label0003.nii.gz
```
### Cache 3d volume into slices
After generating the ```[example]_train.txt``` file, check the config file ```configs/vit_b.yaml```.
Update the params in ```dataset``` by yours. And the ```dataset_list``` should be the name of the generated txt file ```[example]```.
Then run
```
python -m datasets.cache_dataset3d
```
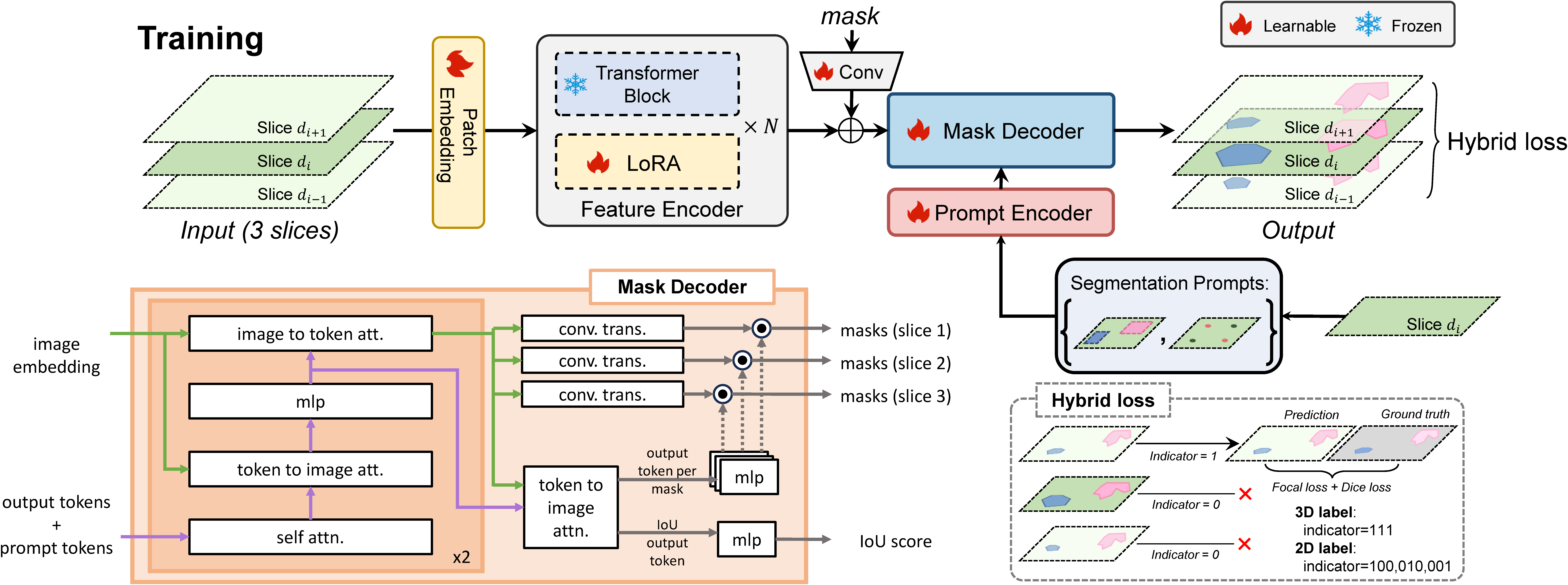
## Start Training
Run training on multi-GPU
```
CUDA_VISIBLE_DEVICES=0,1,2,3 python -m core.ddp --tag debug
```
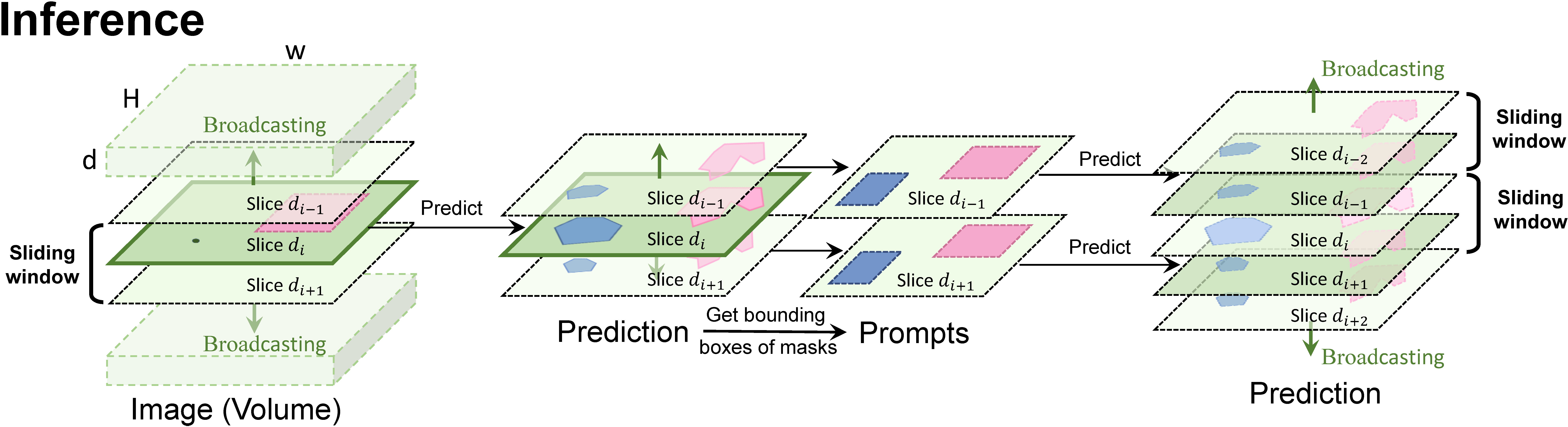
## Sliding Inference and Test
```
python -m core.volume_predictor
```
## TODOs
- [x] Paper released
- [x] Code released
- [x] Slide-SAM-B weights released
- [x] Slide-SAM-H weights released
## Models
### Large scale Medical Image Pretrained Weights
| name | resolution | Prompt | Weights |
| :---------: | :---------: | :---------: | :----------------------------------------------------------: |
| Slide-SAM-B | 1024 x 1024 | box & point | [ckpt](https://pan.baidu.com/s/1jvJ2W4MK24JdpZLwPqMIfA) [code: 7be9] |
| Slide-SAM-H | 1024 x 1024 | box & point | [ckpt](https://pan.baidu.com/s/1jnOwyWd-M1fBIauNi3IA4w) [code: 05dy] |
## Getting Started
### Install tutils tools
```
pip install trans-utils
```
### Prepare datasets
We recommend you to convert the dataset into the nnUNet format.
```
00_custom_dataset
imagesTr
xxx_0000.nii.gz
...
labelsTr
xxx.nii.gz
...
```
Try to use the function organize in [nnunet-style](https://github.com/MIC-DKFZ/nnUNet/blob/master/documentation/dataset_format.md) or ```organize_by_names``` to prepare your custom datasets.
Then run :
```python
python -m datasets.generate_txt
```
A ```[example]_train.txt``` will be generated in ```./datasets/dataset_list/```
The content should be like below
```
01_BCV-Abdomen/Training/img/img0001.nii.gz 01_BCV-Abdomen/Training/label/label0001.nii.gz
01_BCV-Abdomen/Training/img/img0002.nii.gz 01_BCV-Abdomen/Training/label/label0002.nii.gz
01_BCV-Abdomen/Training/img/img0003.nii.gz 01_BCV-Abdomen/Training/label/label0003.nii.gz
```
### Cache 3d volume into slices
After generating the ```[example]_train.txt``` file, check the config file ```configs/vit_b.yaml```.
Update the params in ```dataset``` by yours. And the ```dataset_list``` should be the name of the generated txt file ```[example]```.
Then run
```
python -m datasets.cache_dataset3d
```
## Start Training
Run training on multi-GPU
```
CUDA_VISIBLE_DEVICES=0,1,2,3 python -m core.ddp --tag debug
```
## Sliding Inference and Test
```
python -m core.volume_predictor
```